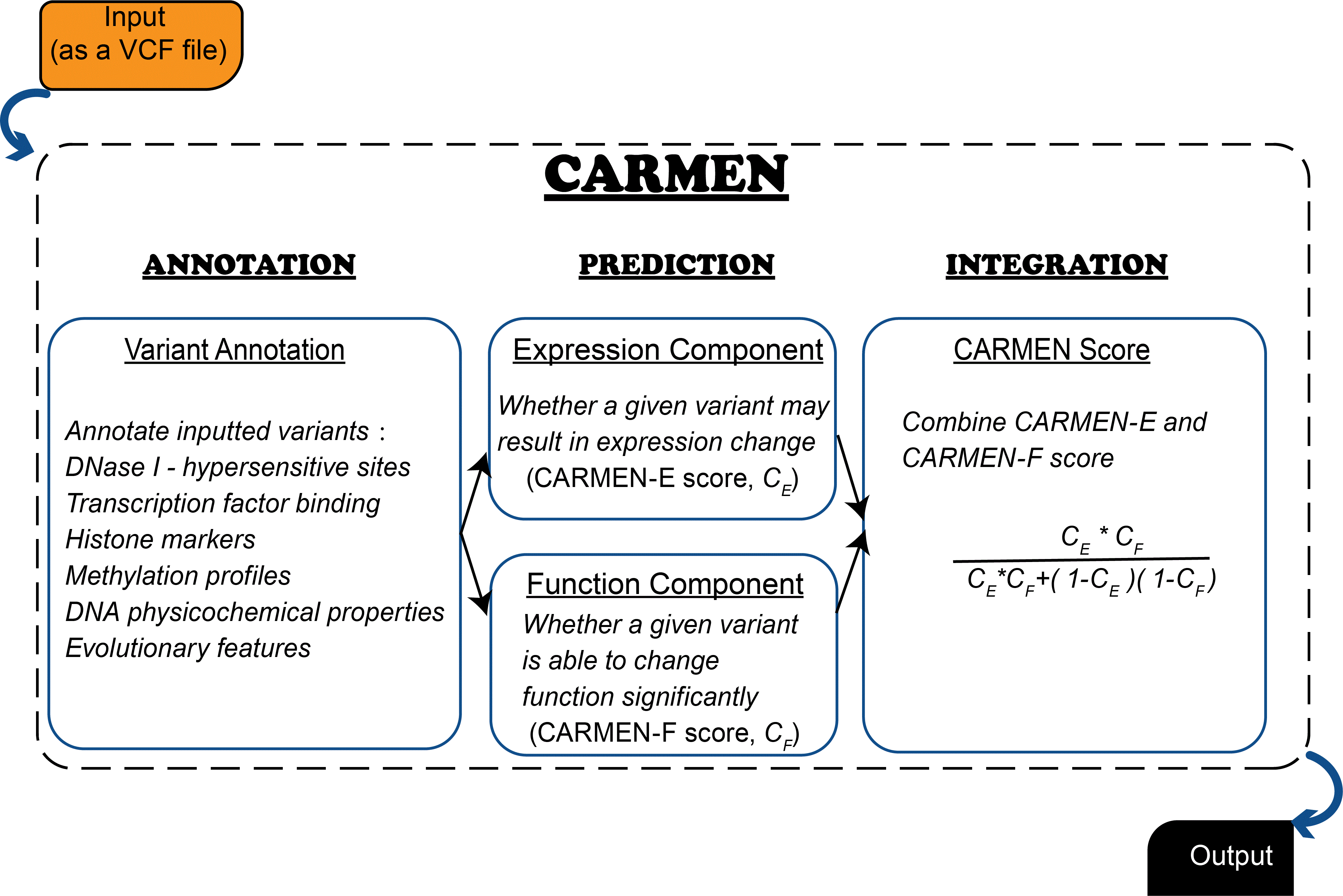
What is CARMEN
Large-scale genome-wide association and expression quantitative trait loci studies have identified multiple noncoding variants associated with genetic diseases via affecting gene expression. However, pinpointing the causal variants effectively and efficiently is still elusive. Here, we developed CARMEN, a novel algorithm to identify functional noncoding expression-modulating variants. Multiple evaluations demonstrate that CARMEN shows superior performance over state-of-the-art tools, and is able to pinpoint potentially causal variants other than lead SNPs reported by association studies. Meanwhile, benefitting from extensive annotations generated, CARMEN provides mechanism hints on predicted expression-modulating variants, enabling effectively characterizing functional variants involved in gene expression and disease-related phenotypes. CARMEN is well scale with massive dataset and is available online as a Web server at http://carmen.gao-lab.org.